What is SOTNIR?
SOTNIR stands for Southampton Neuroimaging Repository.
Purpose
The purpose of establishing SOTNIR is to make it easier to carry out high-quality neuroimaging research at Southampton. This is an initiative between UoS Faculty of Medicine and UHS Imaging Physics Group, but it is important to note that the ever-growing Southampton neuroimaging community spans multiple faculties and disciplines across the UoS and the UHS, therefore, having an infrastructure that allows the storage, management, and sharing of neuroimaging data, software, and protocols represents a genuine priority. SOTNIR aims to be a collaborative platform to enable this, where on-going communications are taking place between scientists and engineers who work in academic or clinical settings. In addition, SOTNIR advocates sharing neuroimage processing software that are well developed, tested, and documented, so that the risks related to clinical applications are well managed, and people coming from different backgrounds know how to use them.
Benefits
The benefit of SOTNIR is multi-folds. Locally, researchers can increase the uptake of projects for publication and funding applications, students can access standardized workflows to carry out short-term projects and fulfill curriculum. Globally, SOTNIR fosters positive interactions with external collaborators and the broader scientific community, enabling re-analysis and meta-analysis of the data in different contexts1, thus presenting a collective effort towards transparent and reproducible science. Many neuroimaging repositories have been made publicly available2–8, covering a wide spectrum of imaging modality, population, and study design. Therefore, establishing SOTNIR is not innovative, but fundamental, for enabling Southampton neuroimaging research to continue expanding, a commitment that the UoS and the UHS has made significant investment.
Components
An illustrative diagram of SOTNIR is provided below (Figure 1):
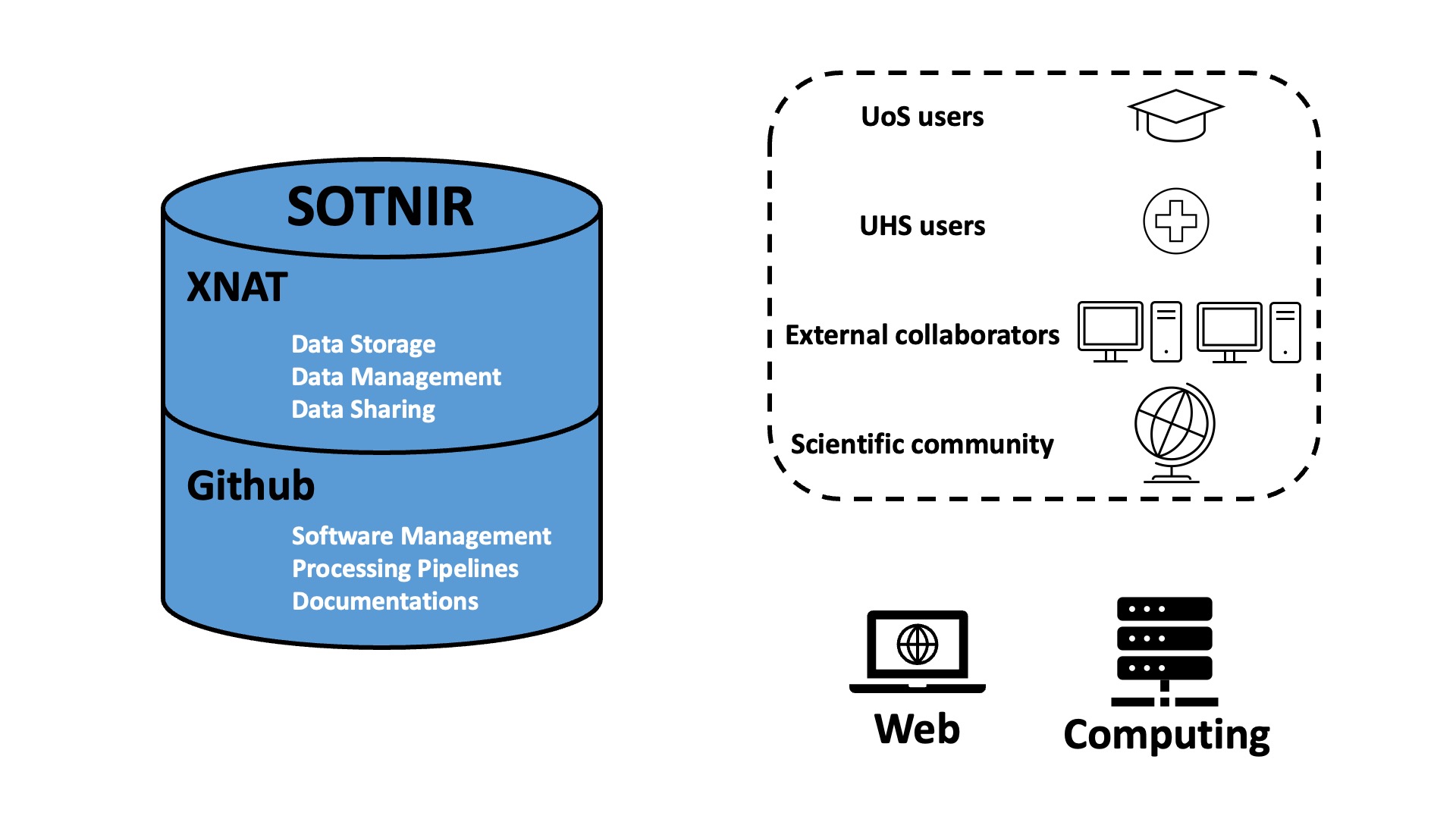
In brief, SOTNIR consists of two components: one that hosts data, and the other that hosts software and protocols:
- eXtensible Neuroimaging Archive Toolkit (XNAT) is a platform that can store, manage, and share neuroimaging data. XNAT has been widely implemented for different projects around the world, such as NITRC2, VUIIS3, NUNDA4, ConnectomeDB5, SchizConnect7, and the Dementia Platform UK9.
- GitHub primarily hosts and provides version control of source code, and GitHub is a “social coding platform” where software developers and researchers can exchange ideas and collaborate on codes10. GitHub allows automated testing, so that new codes can be continuously integrated with existing codes to be well maintained10. GitHub has been extensively used in neuroimaging community, an example is the Diffusion Imaging in Python (DIPY)11. DIPY uses a “public code review” model to maintain high-quality code, where everyone is given a chance to comment and raise questions on proposed contribution before the new code is merged11.
Users
Individuals that interact with SOTNIR can be grouped into four types:
- UoS users
- UHS users
- External collaborators
- General users from the broader scientific community
Routes
There are two routes that individuals may choose from to interact with SOTNIR:
- Web user interface: A subdomain can be applied to host a SOTNIR website at UoS. The website provides a user-friendly interface, where users can navigate and perform query, e.g. search for datasets of certain modality and format (see NITRC2). In addition, the website can post education resources, such as courses on campus and online workshops. Opportunities for doing research, and research studies that are looking for participants can also be posted on the website.
- High-performance computing resources: This usually has a command-line interface. An example would be Iridis 5, a supercomputer housed at UoS. Iridis 5 has more than 20,000 processor cores and a peak of over 1 Petaflop/s, along with 2.5Pb of online storage and 10Pb of deep offline storage. Bespoke software and pipelines can be cloned from SOTNIR to Iridis 5 to process and analyze datasets.
Implementation
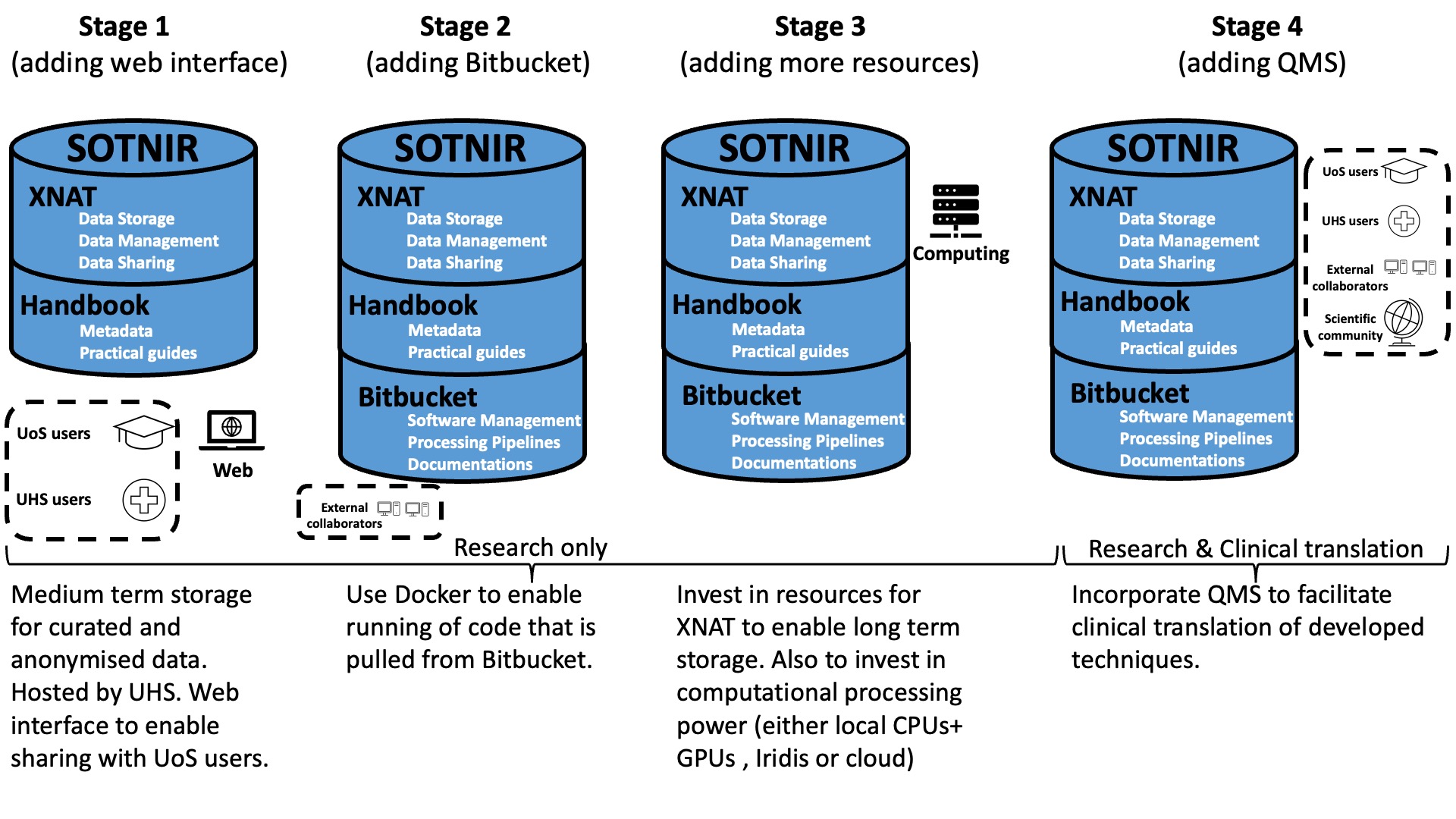
A phased implementation of SOTNIR is illustrated below (Figure 2):